Kudzu density functions and non-parametric Bayesian updating
This page explains my work on this topic and is a repository for code and extended outputs that could not fit into publications. The code is ©BayesCamp Ltd 2019-2024, and licensed for use CC-BY-4.0. The text and images in this web page are also ©BayesCamp Ltd 2019-2024 — except the photograph (Wikimedia) and the flower vector art (dreamstime) — and are NOT licensed for reuse (all rights reserved). You may use the code as you wish, as long as you acknowledge the source. I suggest you publicly display this: "Adapted from code (c) BayesCamp Ltd, Winchester, UK, under the CC-BY-4.0 license".

Primer
Bayesian updating is the principle that you can add data sequentially in batches to learn a posterior distribution. Yesterday's posterior distribution becomes today's prior distribution, and you only need to calculate the likelihood for the current batch of data. If done correctly, the result will be the same as if you had analysed all data together.
In my experience, teachers of intro Bayes courses talk about this all the time. Then it never gets used in real life. Why not? Parametric updating is possible if you correctly know the joint distribution over all the unknowns. You can specify hyperparameters for the next prior by analysing the previous posterior. In the case of a multivariate normal prior and posterior for p unknowns, this would mean a vector of p means and a p-by-p covariance matrix.
But we do not know the distribution. That's why we're doing the analysis. We don't know if it is multivariate normal, any other off-the-shelf distribution, or shaped like nothing we've ever seen before.
A non-parametric updating method would use yesterday's posterior sample to create today's prior density function. In this sense, we have yesterday's posterior sample, and we are going to use it as today's prior sample. Because these samples can have a dual purpose, I call them p[oste]riors (nobody else does, for obvious reasons).
To be totally clear though, the prior that we supply to any Bayesian software is a density function: it receives a vector of unknowns (a point in parameter space), and some hyperparameters, and returns a probability density. This is an application of the general statistical task called density estimation. See David Scott's book, Multivariate Density Estimation (2015) for a recent, clearly-written overview.
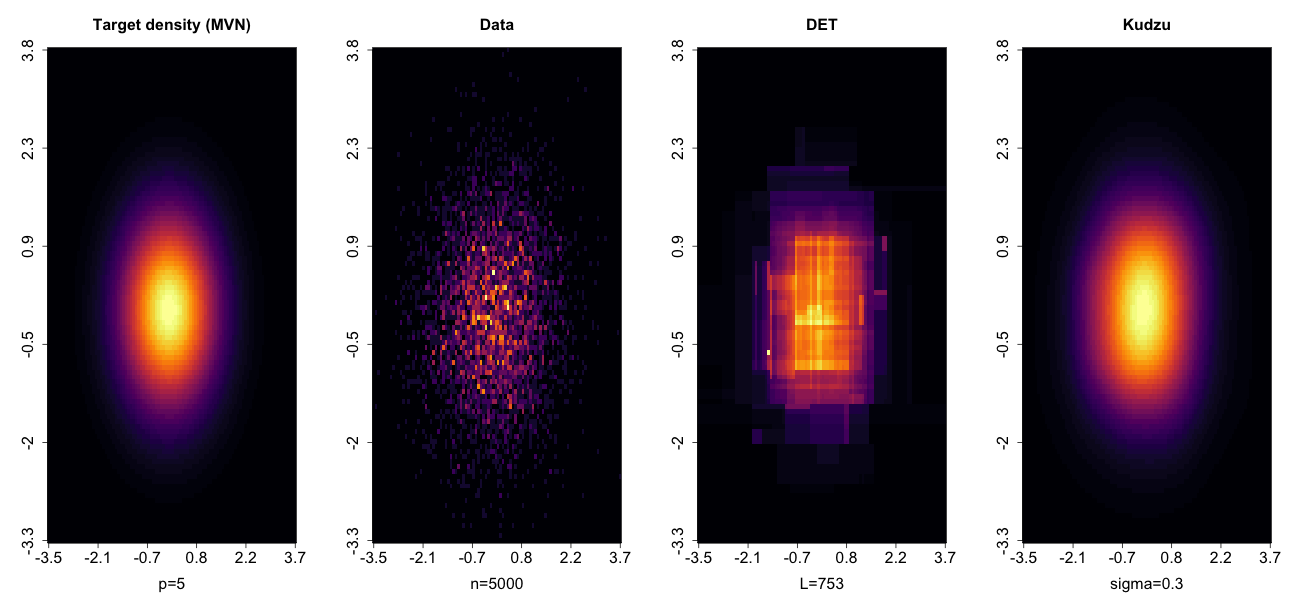
The classic density estimation method is kernel density estimation, but this will not work for anything but the smallest number of dimensions (p), maybe p<5. This is long established and I demonstrated it in this Bayesian updating context in my 2022 IJCEE paper (post-script PDF link available via peer-reviewed paper number 77 in my Publications page).
"Year after year
Feeding cherry trees
Blossom fragments"
— Matsuo Basho (1691), trans. Takafumi Saito & William R Nelson
My alternative proposal is to use density estimation trees (DET), a fast and scalable method which uses the same tree-growing algorithm from classification and regression trees (CART, Breiman et al), but to estimate a density instead of a proportion or a mean. The DET estimate cuts the area the data occupy into hypercuboids called "leaves". The density is then piecewise constant, which will not help with Bayesian sampler algorithms such as Hamiltonian Monte Carlo (implemented in Stan and PyMC*), Metropolis-adjusted Langevin Algorithm or Piecewise Deterministic Markov Processes; these samplers need to obtain and use gradients of the density (the Jacobian vector) and the estimated density should be smooth.
To smooth the DET estimate, I replace the sharp edges of leaves with inverse logistic ramp functions. If you come from a machine learning or signal processing background, you might prefer to think of this as a convolution of each leaf with the logistic probability density function.
I call this the kudzu density function, named after the kudzu vine (Japanese arrowroot), which is an extremely fast-growing plant that grows over, and smooths out, the shapes of trees. All the important facts of kudzu density function work to date is in the pre-print paper in development, which you can download here.

A winter scene of kudzu growing over bushes and trees at Floyd Bennett Field, New York. Image by wikmedia user "Rhododendrites", CC BY-SA 4.0, via Wikimedia Commons
Current pre-print paper
All the important facts of kudzu density function work to date is in the pre-print paper in development, which you can download here. This will continue to expand as work continues through 2024, so check back for the latest.
Previously, early work on this appeared in a 2022 paper in the International Journal of Computational Economics and Econometrics, though the emphasis there is in weighing up variations on kernel density estimates, sampling importance resampling, and kudzu density functions. If you really want to see the historic supporting information for this (and I recommend you just use the current pre-print to learn about it), it is here. Prior to that, I gave a talk on it at the International WebWorkshop on Computational Economics and Econometrics in 2020, hosted by CNR-IRCRES in Rome, Italy but conducted online because of the pandemic (the 2022 paper was in a special issue of talks from that workshop). Even earlier, I had a poster at StanCon in Cambridge, UK, in 2018, although that was all about kernels.
Software for kudzu density functions
Currently, in 2024, I have an R package and Stata commands in progress. The R package is available for alpha testing. You can:
- Fit a density estimation tree that supplies output amenable to kudzu
- Get the integrated squared error: the loss function for the DET and the kudzu density function, which you can use to select tuning parameters
- Obtain kudzu joint density for a given vector
- Obtain kudzu marginal densities, and tail probabilities
- Output code for Stata (bayesmh evaluators syntax), Stan, or BUGS/JAGS, to include a kudzu prior
- Generate (efficiently) pseudo-random numbers from the kudzu density
By downloading this software, you acknowledge that it is work-in-progress, supplied for community testing only, you agree to the license terms and conditions, and you agree that it is supplied as is (voetstoets) with no guarantee of it doing, or not doing, anything whatever, and you agree that you use it at your own risk.
Click here to agree, and download the R package tarball; you will then need to install it locally with install.packages("add_your_path_here/kudzu_0.1.1.tar.gz", repos = NULL, type = "source")
Please note that the R package is entirely in base R, to facilitate scrutiny, sharing of ideas, and open source good vibes. In due course (2025), I will produce a version with the most intensive bits in C++ via Rcpp, which is likely to provide an order of magnitude speed-up, and it is that version that should be used for serious application. The Stata version is implemented almost entirely in Mata, so is already satisfyingly fast. Python and Julia versions can then follow using the same C++ innards.
I have some ideas to do soon, which appear in the pre-print paper. If you would like to test it and send me some ideas, you can email me as shown in the black navbar above. I no longer use Github, so when it is fully released, the R package will be here, and the Stata will be on SSC.